In recent years, there has been a growing trend among scientific researchers and institutions to move their scientific workflows to public cloud platforms. This shift has been driven by a number of factors, including the increased availability of cloud computing resources that can be easily scaled up or down as needed, the ability to easily collaborate and share data with other researchers, and the desire to reduce the costs and complexity of on-premises data centers.
Running scientific workflows on public cloud platforms has several disavantages as well, such as concerns about security and data privacy, challenges around data transfer and network connectivity, and importantly the cost of data ingress and egress.
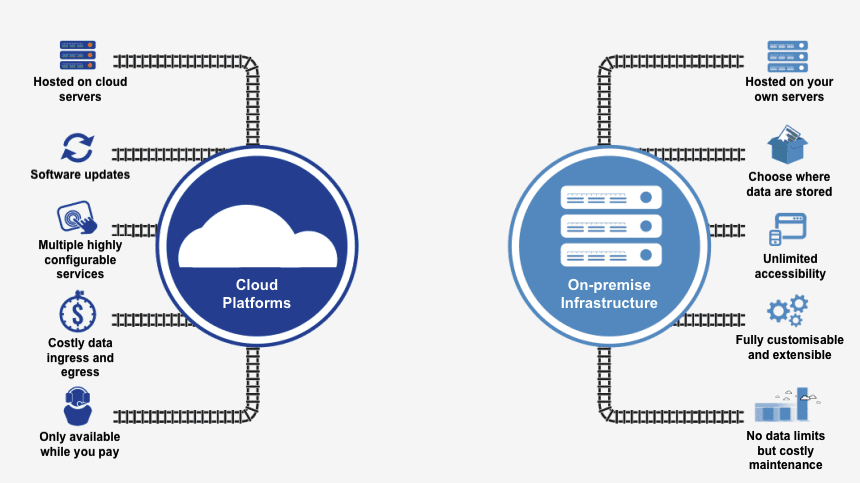
I have recently read two very interesting blog post exactly about this topic: running software and workflows on public cloud versus on-premise. Both articles provide valid but opposing views, which I think are entirely relevant for the discussion, particularly interesting for me in relation to running Bioinformatics workflows.
In one of the articles, the author describes why he thinks we should run software in the cloud2:
- The cloud has significantly changed the way technology is built and has contributed to the evolution of software development in recent years.
- While the cloud has the potential to offer infinite scalability, less time spent on infrastructure, fewer constraints, and lower costs, it has not yet fully delivered on these promises.
- There are still improvements to be made in terms of feedback loops, environment differences, and utilizing new technologies like Docker to their full potential.
- The cloud is still in its early stages and there is potential for further evolution and development, similar to the shifts seen in the music industry with the transition to streaming services.
In the other article, the author describes why he thinks open clould platforms are not cost-effective in particular for running scientific workflows3:
- Generic cloud computing platforms like AWS may not be cost-effective for scientific computing due to their infrastructural complexity and pricing model.
- Scientific computing has different needs than web and mobile apps, including different availability requirements, networking needs, load profiles, and latency tolerance.
- It may be more cost-effective for organizations such as universities, research institutes, and biotech companies to build their own scientific computing infrastructure rather than using a cloud computing platform.
- AWS, Azure, and GCP all have high data egress charges, making it more expensive to transfer large amounts of data out of their platforms.
Running scientific workflows: flexibility is key
One main problem which we need to bring into considerantion, is that a large portion of the scientific work is funded by the public, throught public funding agencies, government grants, etc. The projects are typically funded for a defined period of time and subject to renewal, successful grant applications, and so on. The case for on-premise infrastructure is strong if we think about the life-cycle of most resarch projects.
- What happens when the project runs out of money, or is no longer funded?
- What happens when you can affort to ingest the data and run some computation but the cost of egress is so high that you can no longer exit the platform?
I suppose similar drawbacks can be pointed for on-premise, but generally speaking having infrastructure that you can manage yourself provides ultimate flexility for what and for how long you run the projects and keep data. Managing an IT infrastructure with clear policies for data storage and computation, is very challenging though4.
Conclusion
The trend towards running scientific workflows on public cloud platforms is likely to continue due to its benefits and despite the drawbacks. It is important to carefully consider the costs and benefits of building your own infrastructure versus using a cloud computing platform, as each situation will be unique. Overall, it is an exciting time to be in tech, and the next decade is likely to bring many exciting developments in the field of computing, and in particular considering the current AI advances.
Have you consider moving your workflows to the cloud? or perhaps have done it and are now considering moving them back to local infrastructure? Share your experiences and feedback!
1On-cloud versus on-premises diagram adapted from setplex’s.
2We are still early with the cloud: why software development is overdue for a change by Erik Bernhardsson (Nov 19, 2022)
3AWS doesn’t make sense for scientific computing by Noah Lebovic (Oct 7, 2022)
4Running servers (and services) well is not trivial by Chris Siebenmann (Jun 22, 2018)
