Our latest service update article titled Search and sequence analysis tools services from EMBL-EBI in 2022 has been published in Nucleic Acids Research, Web Server Issue.
In this paper we update the community on the latest developments in both Job Dispatcher sequence analysis services as well as EBI Search. We describe recent improvements to these services and updates made to accommodate the increasing data requirements during the COVID-19 pandemic.
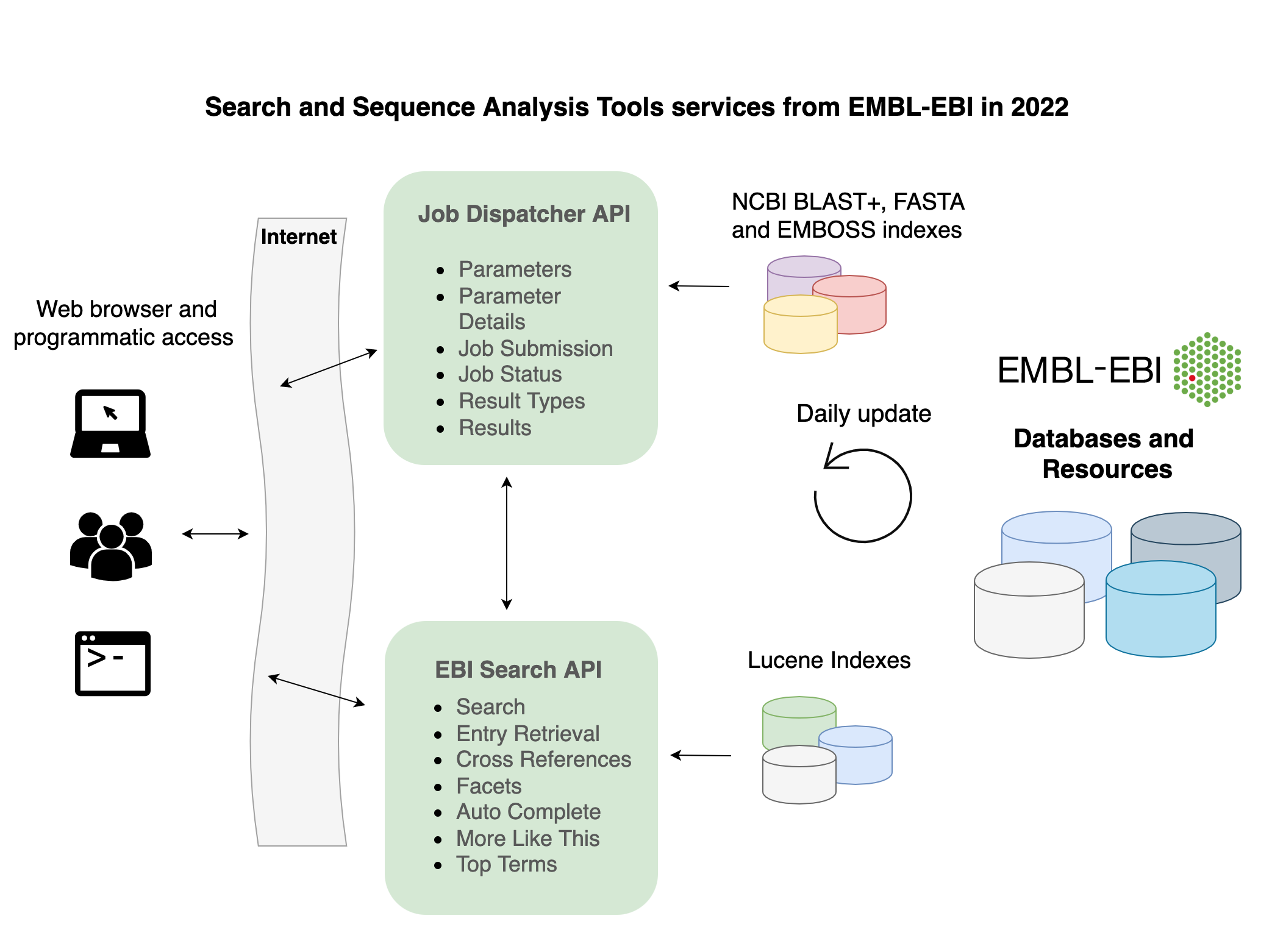
For those not so familiar with Job Dispatcher and EBI Search, the abstract should give a good idea what these are about:
The EMBL-EBI search and sequence analysis tools frameworks provide integrated access to EMBL-EBI’s data resources and core bioinformatics analytical tools. EBI Search (https://www.ebi.ac.uk/ebisearch) provides a full-text search engine across nearly 5 billion entries, while the Job Dispatcher tools framework (https://www.ebi.ac.uk/services) enables the scientific community to perform a diverse range of sequence analysis using popular bioinformatics applications. Both allow users to interact through user-friendly web applications, as well as via RESTful and SOAP-based APIs.
A good summary of all the bioinformatics applications available through JD in 2022 is provided below:
| Category | Tools |
|---|---|
| Multiple Sequence Alignment | Clustal Omega, Kalign, MAFFT, MUSCLE, T-Coffee, MView, WebPrank |
| Pairwise Sequence Alignment | Needle, Stretcher, Water, Matcher, LALIGN, GeneWise, GGSEARCH2SEQ, SSEARCH2SEQ |
| Phylogeny Analysis | Simple Phylogeny |
| Protein Functional Analysis | InterProScan 5, PfamScan, Phobius, Pratt, RADAR, HMMER3 phmmer, HMMER3 hmmscan |
| RNA Analysis | Infernal cmscan, MapMi, R2DT |
| Sequence Similarity Search | NCBI BLAST+, PSI-BLAST, FASTA, SSEARCH, FASTM/S/F, GGSEARCH, GLSEARCH, PSI-Search, PSI-Search2 |
| Sequence Statistics | SAPS, Pepinfo, Pepstats, Pepwindow, Cpgplot, Newcpgreport, Isochore, Dotmatcher, Dottup, Dotpath, Polydot |
| Sequence Translation | Transeq, Sixpack, Backtranseq, Backtranambig |
| Sequence Format Conversion | Seqret, MView |
| Sequence Operation | Seqcksum |
| EMBOSS Suite | Needle, Stretcher, Water, Matcher, Transeq, Sixpack, Backtranseq, Backtranambig, Pepinfo, Pepstats, Pepwindow, Cpgplot, Newcpgreport, Isochore, Dotmatcher, Dottup, Dotpath, Polydot, Seqret |
| Dbfetch | Dbfetch (fetching data from 57 domains) |
Sequence datasets available through JD in 2022 are also provided below:
| Category | Data |
|---|---|
| UniProtKB protein sequences | UniProtKB, SwissProt, SwissProt Isoforms, TrEMBL, UniProtKB Taxonomic Subsets (13 subgroups, including: bacteria, archaea, eukaryota, SARS-CoV-2, etc.), Reference Proteomes, Representative Proteomes (15, 35, 55, 75), UniProt Reference (UniRef 50, 90 and 100), UniParc, Unimes, UniProtKB-PDB |
| Patent protein sequences | EPO, JPO, KIPO, UPSPTO |
| Structures of protein sequences | PDBe, AlphaFold DB |
| Protein families | Pfam, TIGRFAM, Superfamily, Gene3D, PIRSF, TreeFam, Pfam SARS-CoV-2 |
| Other protein sequences | Enzyme Portal, IntAct, IPD-IMGT/HLA, IPD-KIR, IPD-MHC, MEROPS (MP, MPEP and MPRO), ChEMBL, Quest for Orthologs |
| ENA nucleotide sequences | ENA sequences for Coding, Non-coding, Barcode, Geospatial, Ribosomal RNA and others (10 subgroups, including: Expressed Sequence Tag, Genome Survey Sequence, etc.) |
| Ensembl Genomes sequences | Genomes from Bacteria, Fungi, Plants, Metazoa, Protists, WormBase Parasite, SARS-CoV-2 |
| Structures of nucleotide sequences | PDBe |
| Other nucleotide sequences | IMGT/LIGM-DB, IMGT/HLA (CDS and genomic), IPD-KIR (CDS and genomic), IPD-NHKIR (CDS and genomic), IPD-MHC (CDS and genomic) |
| Additional entries available via Dbfetch | EMDB, PDBe-KB, MEDLINE, NCBI Taxonomy, EDAM ontology, HGNC |
This post is a reproduction of the post originally made on the Job Dispatcher Blog
